Workflow Overview
uSort-M transforms a pooled DNA library into an arrayed collection of sequence-verified single clones in approximately one week.
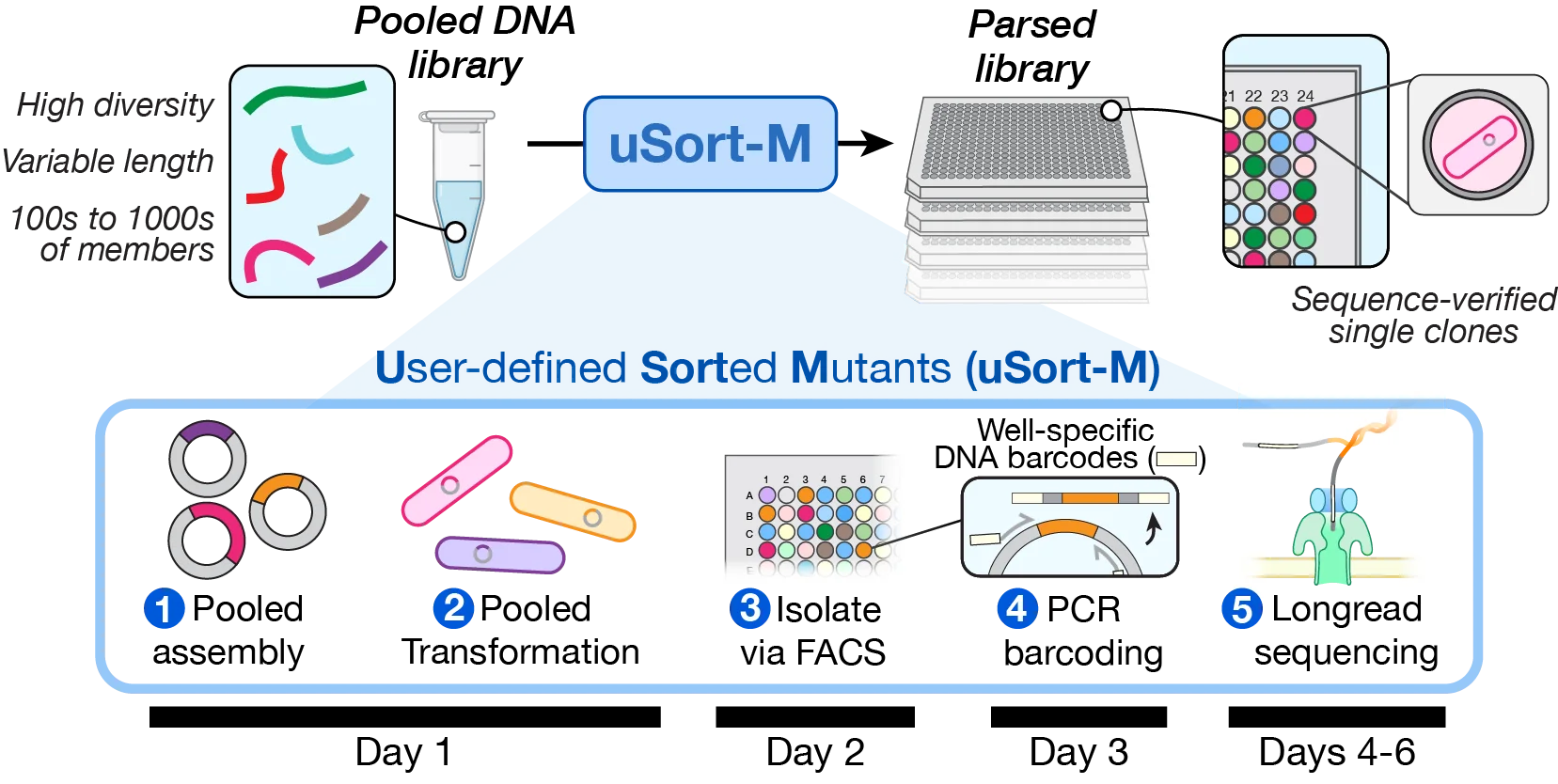
Timeline
| Day | Step | Duration | Notes |
|---|---|---|---|
| 1 | Pooled assembly | 2-3 hours | Golden Gate or Gibson assembly |
| 1 | Transformation | 1-2 hours | Standard chemical transformation |
| 1-2 | Overnight growth | 12-16 hours | Recovery on selective plates |
| 2+ | FACS sorting | ~8 min/plate | Single-cell isolation into 384-well |
| 2+ | Outgrowth | 12-16 hours | Overnight in 384-well plates |
| 3+ | PCR barcoding | ~50 min/plate | Well-specific barcode addition |
| 3+ | Pooling | 1-2 hours | Combine barcoded amplicons |
| 4-6 | Sequencing | 1-3 days | Plasmidsaurus or ONT |
| 6+ | Analysis | 1-2 hours | Demux, variant calling, reporting |
| 6+ | Hit picking | Variable | Automated or manual cherry-picking |
Step-by-Step
1. Library Design
Define your variant library as a list of sequences or mutations. uSort-M works with any pooled DNA source:
- Oligo pools (e.g. Twist Oligo Pools, IDT oPools)
- Error-prone PCR products
- Multiplex assembly libraries
Estimate costs and coverage:
usortm estimate --library-size 500 --seq-length 300 --fold-sampling 4This command calculates projected costs for synthesis, cloning, sorting, barcoding, sequencing, and hit-picking. It also estimates the timeline and effort required for your experiment.
Create an experiment plan:
usortm plan variants.csv --output my_project/ \
--seq-length 300 \
--fold-sampling 4The plan command reads your variant CSV, calculates the number of plates needed, generates barcode assignments, and creates a project directory with a detailed experimental plan.
Learn more about library design →
2. Pooled Assembly
Perform pooled Golden Gate or Gibson assembly to insert your variant sequences into the destination vector.
Key considerations:
- Use a high-fidelity assembly method
- Include a selectable marker
- Target 10-100x transformation coverage
3. FACS Sorting
Use fluorescence-activated cell sorting to isolate single cells into 384-well plates.
Typical parameters:
- Single-cell mode
- FSC/SSC gating for live cells
- ~8 minutes per 384-well plate
- Sort 8-10x library size for good coverage
4. PCR Barcoding
After overnight outgrowth, add well-specific DNA barcodes via PCR.
Recommended barcode sets:
- LevSeq (Arnold lab) - Optimized for Oxford Nanopore
- evSeq - Dual-indexing for Illumina and ONT
5. Sequencing
Pool barcoded amplicons and submit for sequencing.
Recommended platforms:
- Commercial Long-read Sequencing - Plasmidsaurus Custom Sequencing offers affordable multi-Gb library sequencing with fast turnaround
6. Demultiplexing & Analysis
Use the uSort-M CLI to process sequencing data:
Demultiplex sequencing reads:
# Using library CSV (recommended — auto-generates reference FASTA)
usortm demux project/ --fastq data.fastq --library-csv variants.csv
# Or with a pre-built reference FASTA
usortm demux project/ --fastq data.fastq --reference reference.fastaThe demux command runs the LevSeq barcode pipeline: reference alignment with minimap2, strand splitting, barcode demux with Dorado, per-well consensus generation, and variant calling.
Generate pick list:
usortm pick project/Creates an Integra ASSIST PLUS-compatible CSV file for automated cherry-picking of sequence-verified clones, ordered to match your input library.
Generate analysis report:
usortm report project/Produces reports (HTML, CSV, JSON) with coverage statistics, plate maps, and variant recovery metrics.
7. Hit Picking
Cherry-pick sequence-verified clones into a final arrayed format.
The usortm pick command generates Integra ASSIST PLUS-compatible hit lists for automated liquid handling.
File Formats
Variant CSV
Input file listing your library variants. Required columns:
name,sequence
variant_001,ATGGCTAAAGGTGAAGAACTG...
variant_002,ATGGCTAGAGGTGAAGAACTG...
variant_003,ATGGCTAAAGGTGAGGAACTG...Alternative format using mutations (for substitution libraries):
name,mutation
variant_001,A123G
variant_002,T456C
variant_003,G789ABarcode CSV
Well-specific DNA barcodes for demultiplexing. Format:
plate,well,barcode
plate_1,A01,ACGTACGT
plate_1,A02,TGCATGCA
plate_1,A03,GCTAGCTARecommended barcode sets: LevSeq (Arnold lab, ONT-optimized) or evSeq (dual-indexing for Illumina/ONT).
Reference FASTA
Expected sequence for variant calling:
>gene_of_interest
ATGGCTAAAGGTGAAGAACTGTTTACCGGTGTTGTGCCGATTCTGGTGGAACTGGATGGT
GATGTGAACGGTCACAAATTCTCTGTGCGTGGTGAAGGTGAAGGTGATGCTACCTACGGTExpected Results
For a typical 500-variant library with 8x oversampling:
| Metric | Expected Value |
|---|---|
| Wells sorted | ~4,000 |
| Wells with growth | ~2,700 (67%) |
| Unique variants recovered | ~450 (90%) |
| Cost per variant | ~$6-8 |
| Total time | 5-7 days |